|
PLUTO
|
|
PLUTO
|

Go to the source code of this file.
Macros | |
| #define | ARRAY_1D(nx, type) (type *)Array1D(nx,sizeof(type)) |
| #define | ARRAY_2D(nx, ny, type) (type **)Array2D(nx,ny,sizeof(type)) |
| #define | ARRAY_3D(nx, ny, nz, type) (type ***)Array3D(nx,ny,nz,sizeof(type)) |
| #define | ARRAY_4D(nx, ny, nz, nv, type) (type ****)Array4D(nx,ny,nz,nv,sizeof(type)) |
Functions | |
| int | AdvanceStep (const Data *, Riemann_Solver *, Time_Step *, Grid *) |
| void | AdvectFlux (const State_1D *, int, int, Grid *) |
| void | Analysis (const Data *, Grid *) |
| void | Boundary (const Data *, int, Grid *) |
| char * | Array1D (int, size_t) |
| char ** | Array2D (int, int, size_t) |
| char *** | Array3D (int, int, int, size_t) |
| char **** | Array4D (int, int, int, int, size_t) |
| double *** | ArrayBox (long int, long int, long int, long int, long int, long int) |
| double *** | ArrayBoxMap (int, int, int, int, int, int, double *) |
| double *** | ArrayMap (int, int, int, double *) |
| unsigned char *** | ArrayCharMap (int, int, int, unsigned char *) |
| void | BodyForceVectorGet (double **v, double **g, double *, double *, double *, int beg, int end, Grid *grid) |
| void | BodyForcePotentialGet (double **v, double **gphi, double *phi_p, double *phi_c, int beg, int end, Grid *grid) |
| double | BodyForcePotential (double, double, double) |
| void | BodyForceVector (double *, double *, double, double, double) |
| void | ChangeDumpVar () |
| void | CharTracingStep (const State_1D *, int, int, Grid *) |
| void | CheckPrimStates (double **, double **, double **, int, int) |
| int | CheckNaN (double **, int, int, int) |
| int | CloseBinaryFile (FILE *, int) |
| void | ComputeUserVar (const Data *, Grid *) |
| float *** | Convert_dbl2flt (double ***, double, int) |
| void | ConsToPrim3D (Data_Arr, Data_Arr, unsigned char ***, RBox *) |
| void | CreateImage (char *) |
| void | ComputeEntropy (const Data *, Grid *) |
| void | EntropySwitch (const Data *, Grid *) |
| void | EntropyOhmicHeating (const Data *, Data_Arr, double, Grid *) |
| void | FreeArray1D (void *) |
| void | FreeArray2D (void **) |
| void | FreeArray3D (void ***) |
| void | FreeArray4D (void ****) |
| void | FreeArrayBox (double ***, long, long, long) |
| void | FreeArrayBoxMap (double ***, int, int, int, int, int, int) |
| void | FreeArrayMap (double ***) |
| void | FreeArrayCharMap (unsigned char ***) |
| void | FindShock (const Data *, Grid *) |
| void | FlagShock (const Data *, Grid *) |
| void | Flatten (const State_1D *, int, int, Grid *) |
| void | FreeGrid (Grid *) |
| void | GetAreaFlux (const State_1D *, double **, double **, int, int, Grid *) |
| void | GetCGSUnits (double *u) |
| Image * | GetImage (char *) |
| double * | GetInverse_dl (const Grid *) |
| int | GetNghost (void) |
| void | GetOutputFrequency (Output *, const char *) |
| RBox * | GetRBox (int, int) |
| double *** | GetUserVar (char *) |
| void | HancockStep (const State_1D *, int, int, Grid *) |
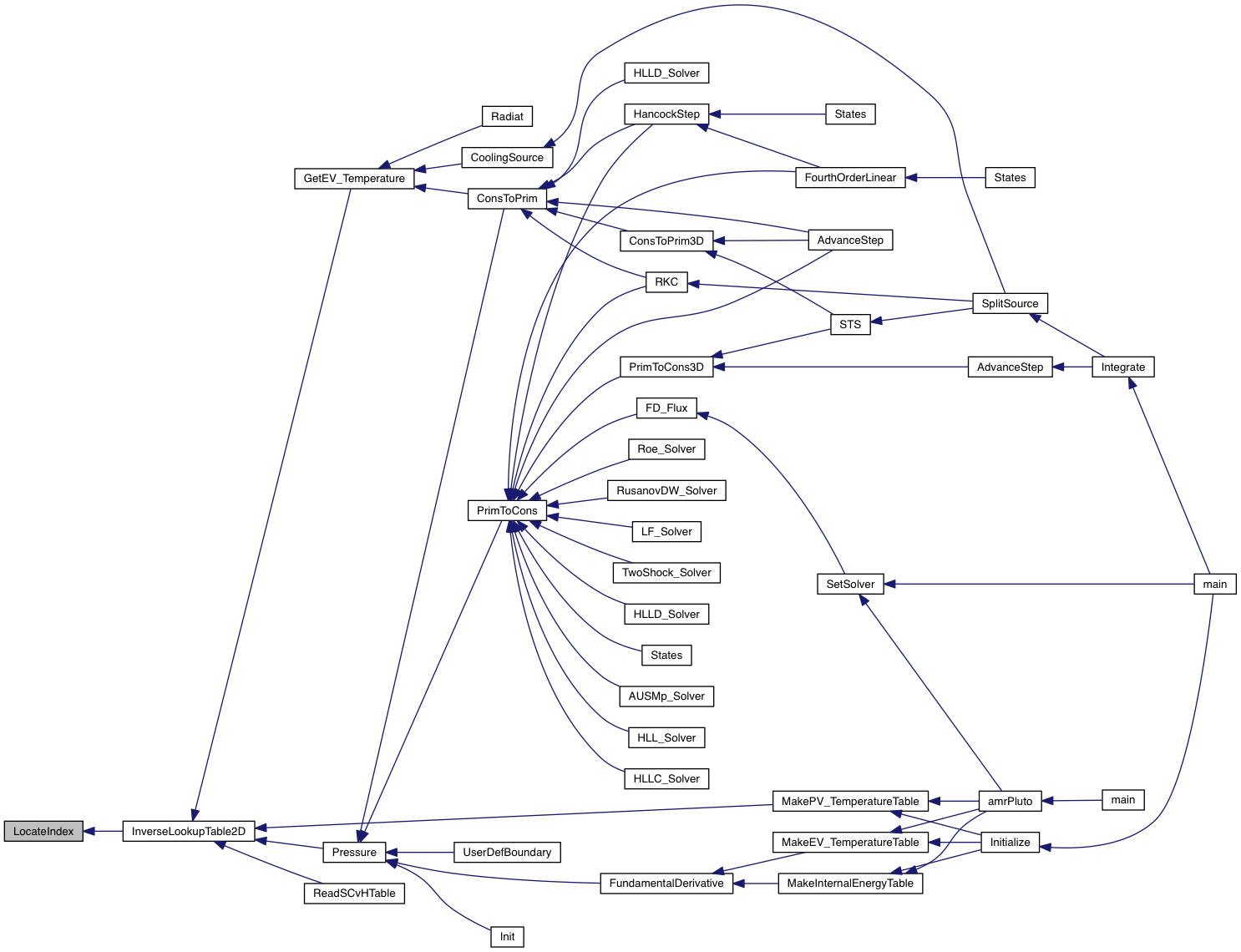
| int | LocateIndex (double *, int, int, double) |
| void | Init (double *, double, double, double) |
| void | Initialize (int argc, char *argv[], Data *, Runtime *, Grid *, Cmd_Line *) |
| void | InternalBoundaryReset (const State_1D *, Time_Step *, int, int, Grid *) |
| void | InputDataFree (void) |
| void | InputDataInterpolate (double *, double, double, double) |
| void | InputDataRead (char *, char *) |
| void | InputDataSet (char *, int *) |
| int | IsLittleEndian (void) |
| void | MakeState (State_1D *) |
| void | MakeGeometry (Grid *) |
| double | MeanMolecularWeight (double *) |
| double | Median (double a, double b, double c) |
| FILE * | OpenBinaryFile (char *, int, char *) |
| void | ParabolicFlux (Data_Arr, Data_Arr J, double ***, const State_1D *, double **, int, int, Grid *) |
| double | ParabolicRHS (const Data *, Data_Arr, double, Grid *) |
| void | ParseCmdLineArgs (int, char *argv[], char *, Cmd_Line *) |
| int | ParamFileRead (char *) |
| char * | ParamFileGet (const char *, int) |
| int | ParamExist (const char *) |
| int | ParamFileHasBoth (const char *, const char *) |
| void | PrimToChar (double **, double *, double *) |
| void | PrimToCons3D (Data_Arr, Data_Arr, RBox *) |
| void | ReadBinaryArray (void *, size_t, int, FILE *, int, int) |
| void | ReadHDF5 (Output *output, Grid *grid) |
| void | ResetState (const Data *, State_1D *, Grid *) |
| void | RestartFromFile (Runtime *, int, int, Grid *) |
| void | RestartDump (Runtime *) |
| void | RestartGet (Runtime *, int, int, int) |
| void | RightHandSide (const State_1D *, Time_Step *, int, int, double, Grid *) |
| void | RightHandSideSource (const State_1D *, Time_Step *, int, int, double, double *, Grid *) |
| void | RKC (const Data *d, Time_Step *, Grid *) |
| Runtime * | RuntimeGet (void) |
| int | RuntimeSetup (Runtime *, Cmd_Line *, char *) |
| void | RuntimeSet (Runtime *runtime) |
| void | SetColorMap (unsigned char *, unsigned char *, unsigned char *, char *) |
| void | SetDefaultVarNames (Output *) |
| int | SetDumpVar (char *, int, int) |
| void | SetIndexes (Index *indx, Grid *grid) |
| int | SetLogFile (char *, Cmd_Line *) |
| void | SetOutput (Data *d, Runtime *input) |
| void | SetRBox (void) |
| Riemann_Solver * | SetSolver (const char *) |
| void | SetGrid (Runtime *, Grid *) |
| void | SetJetDomain (const Data *, int, int, Grid *) |
| void | Show (double **, int) |
| void | ShowMatrix (double **, int n, double) |
| void | ShowVector (double *, int n) |
| void | ShowConfig (int, char *a[], char *) |
| void | ShowDomainDecomposition (int, Grid *) |
| void | ShowUnits () |
| void | SplitSource (const Data *, double, Time_Step *, Grid *) |
| void | STS (const Data *d, Time_Step *, Grid *) |
| void | Startup (Data *, Grid *) |
| void | States (const State_1D *, int, int, Grid *) |
| void | SwapEndian (void *, const int) |
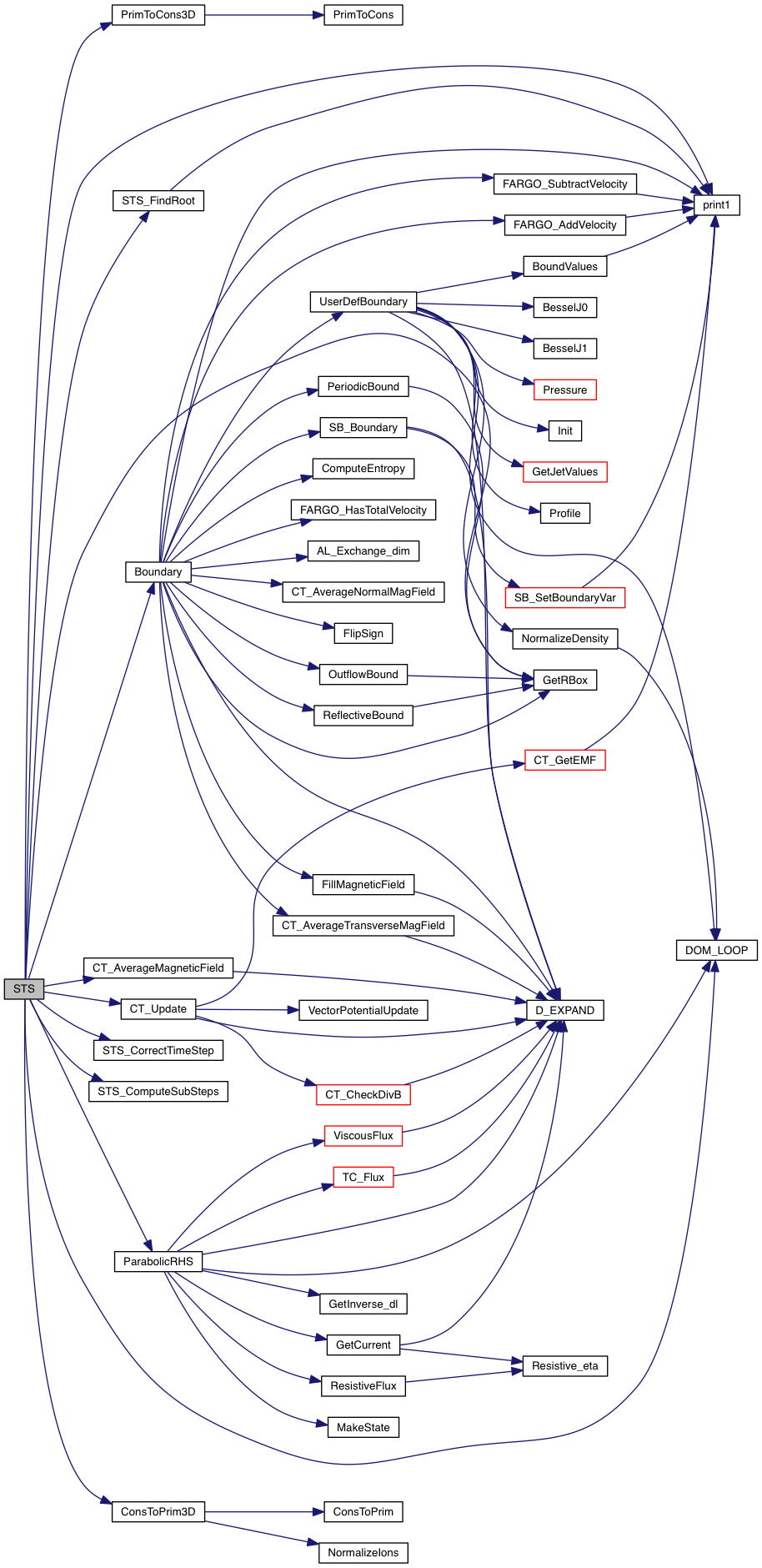
| void | UnsetJetDomain (const Data *, int, Grid *) |
| void | UpdateStage (const Data *, Data_Arr, double **, Riemann_Solver *, double, Time_Step *, Grid *) |
| void | UserDefBoundary (const Data *, RBox *, int, Grid *) |
| void | VectorPotentialDiff (double *, int, int, int, Grid *) |
| void | Where (int, Grid *) |
| void | WriteData (const Data *, Output *, Grid *) |
| void | WriteBinaryArray (void *, size_t, int, FILE *, int) |
| void | WriteHDF5 (Output *output, Grid *grid) |
| void | WriteVTK_Header (FILE *, Grid *) |
| void | WriteVTK_Vector (FILE *, Data_Arr, double, char *, Grid *) |
| void | WriteVTK_Scalar (FILE *, double ***, double, char *, Grid *) |
| void | WriteTabArray (Output *, char *, Grid *) |
| void | WritePPM (double ***, char *, char *, Grid *) |
| void | WritePNG (double ***, char *, char *, Grid *) |
| void | print (const char *fmt,...) |
| void | print1 (const char *fmt,...) |
| void | Trace (double) |
| void | FlipSign (int, int, int *) |
| void | OutflowBound (double ***, int, int, Grid *) |
| void | PeriodicBound (double ***, int, int) |
| void | ReflectiveBound (double ***, int, int, int) |
| void | PlutoError (int, char *) |
| double | Length_1 (int i, int j, int k, Grid *) |
| double | Length_2 (int i, int j, int k, Grid *) |
| double | Length_3 (int i, int j, int k, Grid *) |
| void | VectorPotentialUpdate (const Data *d, const void *vp, const State_1D *state, const Grid *grid) |
| void | Enthalpy (double **, double *, int, int) |
| void | Entropy (double **, double *, int, int) |
| void | SoundSpeed2 (double **, double *, double *, int, int, int, Grid *) |
| double | GetEntropy (double x) |
| void | WriteAsciiFile (char *fname, double *q, int nvar) |
| #define ARRAY_1D | ( | nx, | |
| type | |||
| ) | (type *)Array1D(nx,sizeof(type)) |
Definition at line 170 of file prototypes.h.
| #define ARRAY_2D | ( | nx, | |
| ny, | |||
| type | |||
| ) | (type **)Array2D(nx,ny,sizeof(type)) |
Definition at line 171 of file prototypes.h.
| #define ARRAY_3D | ( | nx, | |
| ny, | |||
| nz, | |||
| type | |||
| ) | (type ***)Array3D(nx,ny,nz,sizeof(type)) |
Definition at line 172 of file prototypes.h.
| #define ARRAY_4D | ( | nx, | |
| ny, | |||
| nz, | |||
| nv, | |||
| type | |||
| ) | (type ****)Array4D(nx,ny,nz,nv,sizeof(type)) |
Definition at line 173 of file prototypes.h.
| int AdvanceStep | ( | const Data * | d, |
| Riemann_Solver * | Riemann, | ||
| Time_Step * | Dts, | ||
| Grid * | grid | ||
| ) |
Advance equations using the corner transport upwind method (CTU)
| [in,out] | d | pointer to Data structure |
| [in] | Riemann | pointer to a Riemann solver function |
| [in,out] | Dts | pointer to time step structure |
| [in] | grid | pointer to array of Grid structures |
Advance the equations by a single time step using unsplit integrators based on the method of lines.
| [in,out] | d | pointer to Data structure |
| [in] | Riemann | pointer to a Riemann solver function |
| [in,out] | Dts | pointer to time step structure |
| [in] | grid | pointer to array of Grid structures |
Definition at line 88 of file ctu_step.c.
| [in,out] | state | |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
Definition at line 47 of file adv_flux.c.
Perform runtime data analysis.
| [in] | d | the PLUTO Data structure |
| [in] | grid | pointer to array of Grid structures |
Compute volume-integrated magnetic pressure, Maxwell and Reynolds stresses. Save them to "averages.dat"
Definition at line 66 of file init.c.
| char* Array1D | ( | int | nx, |
| size_t | dsize | ||
| ) |
Allocate memory for a 1-D array of any basic data type starting at 0.
| [in] | nx | number of elements to be allocated |
| [in] | dsize | data-type of the array to be allocated |
Definition at line 80 of file arrays.c.

| char** Array2D | ( | int | nx, |
| int | ny, | ||
| size_t | dsize | ||
| ) |
Allocate memory for a 2-D array of any basic data type.
| [in] | nx | number of elements in the 2nd dimension |
| [in] | ny | number of elements in the 1st dimension |
| [in] | dsize | data-type of the array to be allocated |
Definition at line 108 of file arrays.c.

| char*** Array3D | ( | int | nx, |
| int | ny, | ||
| int | nz, | ||
| size_t | dsize | ||
| ) |
Allocate memory for a 3-D array of any basic data type.
| [in] | nx | number of elements in the 3rd dimension |
| [in] | ny | number of elements in the 2nd dimension |
| [in] | nz | number of elements in the 1st dimension |
| [in] | dsize | data-type of the array to be allocated |
Definition at line 149 of file arrays.c.
| char**** Array4D | ( | int | nx, |
| int | ny, | ||
| int | nz, | ||
| int | nv, | ||
| size_t | dsize | ||
| ) |
Allocate memory for a 4-D array of any basic data type.
| [in] | nx | number of elements in the 4th dimension |
| [in] | ny | number of elements in the 3rd dimension |
| [in] | nz | number of elements in the 2nd dimension |
| [in] | nv | number of elements in the 1st dimension |
| [in] | dsize | data-type of the array to be allocated |
Definition at line 222 of file arrays.c.

| double*** ArrayBox | ( | long int | nrl, |
| long int | nrh, | ||
| long int | ncl, | ||
| long int | nch, | ||
| long int | ndl, | ||
| long int | ndh | ||
| ) |
Allocate memory for a 3-D array in double precision with given subscript range [low...high] for each direction. Useful for staggered arrays which do not start at [0].
| [in] | nrl | lower bound index for the 3rd dimension |
| [in] | nrh | upper bound index for the 3rd dimension |
| [in] | ncl | lower bound index for the 2nd dimension |
| [in] | nch | upper bound index for the 2nd dimension |
| [in] | ndl | lower bound index for the 1st dimension |
| [in] | ndh | upper bound index for the 1st dimension |
Definition at line 341 of file arrays.c.
| double*** ArrayBoxMap | ( | int | nrl, |
| int | nrh, | ||
| int | ncl, | ||
| int | nch, | ||
| int | ndl, | ||
| int | ndh, | ||
| double * | uptr | ||
| ) |
| unsigned char*** ArrayCharMap | ( | int | , |
| int | , | ||
| int | , | ||
| unsigned char * | |||
| ) |
| double*** ArrayMap | ( | int | nx, |
| int | ny, | ||
| int | nz, | ||
| double * | uptr | ||
| ) |
Convert a one dimensional array with (nx*ny*nz) elements into a 3D array with index range [0..nx-1][0..ny-1][0..nz-1]. This function is similar, conceptually, to Array3D() except that the memory area is already allocated.
| [in] | uptr | pointer to 1D array |
| [in] | nx | number of elements in the 3rd dimension |
| [in] | ny | number of elements in the 2nd dimensions |
| [in] | nz | number of elements in the 1st dimensions |
Definition at line 421 of file arrays.c.
| double BodyForcePotential | ( | double | , |
| double | , | ||
| double | |||
| ) |
| void BodyForcePotentialGet | ( | double ** | v, |
| double ** | gphi, | ||
| double * | phi_p, | ||
| double * | phi_c, | ||
| int | beg, | ||
| int | end, | ||
| Grid * | grid | ||
| ) |
| void BodyForceVector | ( | double * | v, |
| double * | g, | ||
| double | x, | ||
| double | y, | ||
| double | z | ||
| ) |
| void BodyForceVectorGet | ( | double ** | v, |
| double ** | g, | ||
| double * | , | ||
| double * | , | ||
| double * | , | ||
| int | beg, | ||
| int | end, | ||
| Grid * | grid | ||
| ) |
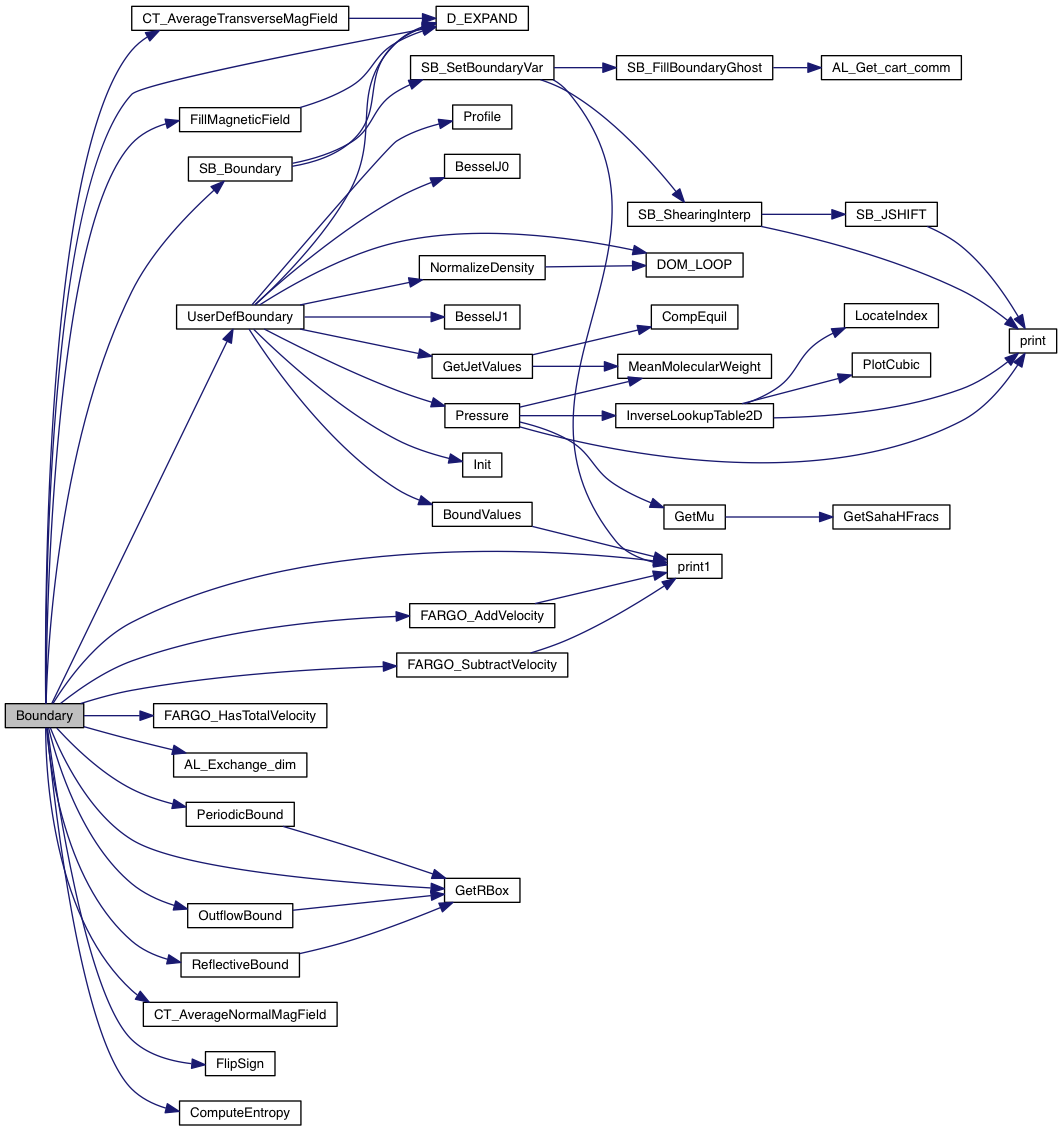
Set boundary conditions on one or more sides of the computational domain.
| [in,out] | d | pointer to PLUTO Data structure containing the solution array (including centered and staggered fields) |
| [in] | idim | specifies on which side(s) of the computational domain boundary conditions must be set. Possible values are
|
| [in] | grid | pointer to an array of grid structures. |
Definition at line 36 of file boundary.c.
| void ChangeDumpVar | ( | ) |
Definition at line 21 of file userdef_output.c.


Compute interface states using characteristic tracing step.
| [in] | state | pointer to a State_1D structure |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
| [in] | grid | pointer to an array of Grid structures |
Tasks are numbered below.
1) Compute eigenvectors and eigenvalues if not yet defined.
2) Obtain characteristic variable increments dwp and dwm.
3) Initialize vp and vm to the reference state. Since this is somewhat arbitrary we use the value of CHTR_REF_STATE to select one of the following cases:
Passive scalars use always CHTR_REF_STATE == 2.
4) Compute left and right states in primitive variables. This step also depends on the value of CHTR_REF_STATE and include:
5) Add source term to L/R states
6) Repeat construction for passive scalars
Definition at line 198 of file char_tracing.c.

| int CheckNaN | ( | double ** | u, |
| int | is, | ||
| int | ie, | ||
| int | id | ||
| ) |
| void CheckPrimStates | ( | double ** | , |
| double ** | , | ||
| double ** | , | ||
| int | , | ||
| int | |||
| ) |
| int CloseBinaryFile | ( | FILE * | fbin, |
| int | sz | ||
| ) |
Close file.
| [in] | fbin | pointer to the FILE that needs to be closed (serial mode only) |
| [in] | sz | the distributed array descriptor for parallel mode |
Definition at line 78 of file bin_io.c.

Definition at line 4 of file userdef_output.c.

Convert a 3D array of conservative variables U to an array of primitive variables V. Note that [nv] is the fastest running index for U while it is the slowest running index for V.
| [in] | U | pointer to 3D array of conserved variables, with array indexing [k][j][i][nv] |
| [out] | V | pointer to 3D array of primitive variables, with array indexing [nv][k][j][i] |
| [in,out] | flag | pointer to 3D array of flags. |
| [in] | box | pointer to RBox structure containing the domain portion over which conversion must be performed. |
Definition at line 16 of file mappers3D.c.
| float*** Convert_dbl2flt | ( | double *** | Vdbl, |
| double | unit, | ||
| int | swap_endian | ||
| ) |
Convert the a double-precision 3D array into single precision. Swap endianity if swap_endian == 1.
| [in] | Vdbl | pointer to a 3D double precision aray |
| [in] | unit | a multiplicative constant typically used to write in c.g.s units. |
| [in] | swap_endian | when set to 1, swap endianity during the conversion. |
Definition at line 216 of file bin_io.c.
| void CreateImage | ( | char * | ) |
| void Enthalpy | ( | double ** | v, |
| real * | h, | ||
| int | beg, | ||
| int | end | ||
| ) |
Compute the enthalpy.
| [in] | v | 1D array of primitive quantities |
| [in] | h | 1D array of enthalpy values |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
Definition at line 48 of file eos.c.
| void Entropy | ( | double ** | v, |
| double * | s, | ||
| int | beg, | ||
| int | end | ||
| ) |
Compute the entropy.
| [in] | v | 1D array of primitive quantities |
| [in] | s | 1D array of entropy values |
| [in] | is | initial index of computation |
| [in] | ie | final index of computation |
Definition at line 80 of file eos.c.

Definition at line 4 of file flatten.c.
| void FlipSign | ( | int | side, |
| int | type, | ||
| int * | vsign | ||
| ) |
Reverse the sign of vector components with respect to axis side. Depending on type, one needs to symmetrize or anti-symmetrize:
where (n) is the normal components, (p) and (t) are the transverse (or poloidal and toroidal for cylindrical and spherical coordinates) components.
| [in] | side | boundary side |
| [in] | type | boundary condition type |
| [out] | vsign | an array of values (+1 or -1) giving the sign |
Definition at line 408 of file boundary.c.
| void FreeArray1D | ( | void * | v | ) |
| void FreeArray2D | ( | void ** | m | ) |
| void FreeArray3D | ( | void *** | m | ) |
| void FreeArray4D | ( | void **** | m | ) |
| void FreeArrayBox | ( | double *** | t, |
| long | nrl, | ||
| long | ncl, | ||
| long | ndl | ||
| ) |
Free memory allocated by the ArrayBox function.
| [in] | t | pointer to an allocated memory area |
| [in] | nrl | starting index of the array for the 3rd dimension |
| [in] | ncl | starting index of the array for the 2nd dimension |
| [in] | ndl | starting index of the array for the 1st dimension |
Definition at line 403 of file arrays.c.

| void FreeArrayBoxMap | ( | double *** | t, |
| int | nrl, | ||
| int | nrh, | ||
| int | ncl, | ||
| int | nch, | ||
| int | ndl, | ||
| int | ndh | ||
| ) |
Free memory allocated with the ArrayBoxMap () function
| void FreeArrayCharMap | ( | unsigned char *** | ) |
| void FreeArrayMap | ( | double *** | t | ) |
Free memory allocate with ArrayMap()
| void FreeGrid | ( | Grid * | grid | ) |
Free array memory allocated previously.
Definition at line 191 of file set_grid.c.
Definition at line 4 of file get_area_flux.c.
| void GetCGSUnits | ( | double * | u | ) |
Compute an array of c.g.s units
| [in] | u | an array containing the c.g.s units of the primitive variables, e.g., u[RHO] will be in gr/cm^3, u[VX1] will be in cm/s, etc... |
Definition at line 557 of file write_data.c.

| double GetEntropy | ( | double | x | ) |
| Image* GetImage | ( | char * | ) |
Definition at line 35 of file set_image.c.
| double* GetInverse_dl | ( | const Grid * | ) |
Definition at line 205 of file set_geometry.c.
| int GetNghost | ( | void | ) |
Compute the number of ghost zones, depending on the selected scheme.
Definition at line 18 of file get_nghost.c.
| void GetOutputFrequency | ( | Output * | output, |
| const char * | output_format | ||
| ) |
Set the intervals between output files. This can be done in three different ways:
However, dn and dclock are mutually exclusive.
Definition at line 433 of file runtime_setup.c.

| RBox* GetRBox | ( | int | side, |
| int | vpos | ||
| ) |
Returns a pointer to a local static RBox
| [in] | side | the region of the computational domain where the box is required. There 8 possible values: X1_BEG, ... , X3_END, DOM, TOT. |
| [in] | vpos | the variable position inside the cell: CENTER, X1FACE, X2FACE or X3FACE. |
Definition at line 232 of file rbox.c.
| double*** GetUserVar | ( | char * | var_name | ) |
return a pointer to the 3D array associated with the variable named 'var_name'.
Definition at line 251 of file set_output.c.
Definition at line 136 of file hancock.c.
| void Init | ( | double * | v, |
| double | x1, | ||
| double | x2, | ||
| double | x3 | ||
| ) |
The Init() function can be used to assign initial conditions as as a function of spatial position.
| [out] | v | a pointer to a vector of primitive variables |
| [in] | x1 | coordinate point in the 1st dimension |
| [in] | x2 | coordinate point in the 2nd dimension |
| [in] | x3 | coordinate point in the 3rdt dimension |
The meaning of x1, x2 and x3 depends on the geometry:
![\[ \begin{array}{cccl} x_1 & x_2 & x_3 & \mathrm{Geometry} \\ \noalign{\medskip} \hline x & y & z & \mathrm{Cartesian} \\ \noalign{\medskip} R & z & - & \mathrm{cylindrical} \\ \noalign{\medskip} R & \phi & z & \mathrm{polar} \\ \noalign{\medskip} r & \theta & \phi & \mathrm{spherical} \end{array} \]](form_173.png)
Variable names are accessed by means of an index v[nv], where nv = RHO is density, nv = PRS is pressure, nv = (VX1, VX2, VX3) are the three components of velocity, and so forth.
Definition at line 17 of file init.c.
| void Initialize | ( | int | argc, |
| char * | argv[], | ||
| Data * | data, | ||
| Runtime * | runtime, | ||
| Grid * | grid, | ||
| Cmd_Line * | cmd_line | ||
| ) |
Initialize computational grid, domain decomposition and memory allocation. Also, set initial conditions and output attributes.
| [in] | argc | the number of command-line argument passed to the code |
| [in] | argv | the argument value as a 1D array of char |
| [in,out] | data | a pointer to the main PLUTO data structure |
| [in,out] | runtime | a pointer to the Runtime structure |
| [in] | grid | pointer to an array of Grid structures |
| [in] | cmd_line | pointer to the Cmd_Line structure |
Definition at line 37 of file initialize.c.

| void InputDataFree | ( | void | ) |
Free memory stored by user-supplied data.
Definition at line 484 of file input_data.c.
| void InputDataInterpolate | ( | double * | vs, |
| double | x1, | ||
| double | x2, | ||
| double | x3 | ||
| ) |
Perform bi- or tri-linear interpolation on external dataset to compute vs[] at the given point {x1,x2,x3}.
| [in] | vs | interpolated value |
| [in] | x1 | coordinate point at which at interpolates are desired |
| [in] | x2 | coordinate point at which at interpolates are desired |
| [in] | x3 | coordinate point at which at interpolates are desired |
The function performs the following tasks.
Definition at line 287 of file input_data.c.

| void InputDataRead | ( | char * | data_fname, |
| char * | endianity | ||
| ) |
Read input data file and store the contents into the local storage array Vin. Memory allocation is also done here. The grid size and number of variables must have previously set by calling InputDataSet().
| [in] | data_fname | the data file name |
| [in] | endianity | an input string ("little" or "big") giving the byte-order of how the input data file was originally written. If an empty string is supplied, no change is made. |
Definition at line 191 of file input_data.c.

| void InputDataSet | ( | char * | grid_fname, |
| int * | get_var | ||
| ) |
Initialize access to input data file by assigning values to grid-related information (geometry, number of points, etc...). This function should be called just once for input-data initialization.
| [in] | gname | the grid file name |
| [in] | get_var | an array of integers specifying which variables have to be read from the input data. |
The following tasks are performed.
Definition at line 54 of file input_data.c.

| int IsLittleEndian | ( | void | ) |
| double Length_1 | ( | int | i, |
| int | j, | ||
| int | k, | ||
| Grid * | |||
| ) |
| double Length_2 | ( | int | i, |
| int | j, | ||
| int | k, | ||
| Grid * | |||
| ) |
| double Length_3 | ( | int | i, |
| int | j, | ||
| int | k, | ||
| Grid * | |||
| ) |
| int LocateIndex | ( | double * | yarr, |
| int | beg, | ||
| int | end, | ||
| double | y | ||
| ) |
Given an array yarr[beg..end] and a given value y, returns the array index i such that yarr[i] < y < yarr[i+1] (for increasing arrays) or yarr[i+1] < y < yarr[i] (for decreasing arrays). yarr must be monotonically increasing or monotonically decreasing.
| [in] | yarr | the 1D array |
| [in] | beg | initial index of the array |
| [in] | end | final index of the array |
| [in] | y | the value to be searched for |
Reference
Definition at line 144 of file math_table2D.c.
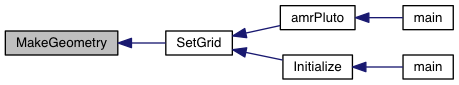
| void MakeGeometry | ( | Grid * | GXYZ | ) |
| [in,out] | GXYZ | Pointer to an array of Grid structures; |
Definition at line 16 of file set_geometry.c.
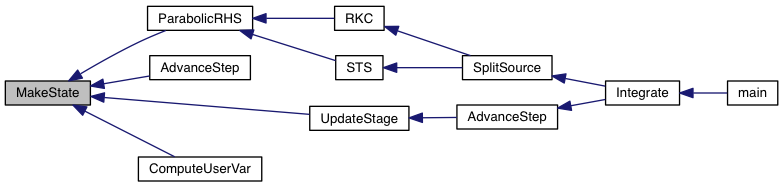
| void MakeState | ( | State_1D * | state | ) |
Allocate memory areas for arrays inside the state structure.
Definition at line 51 of file tools.c.
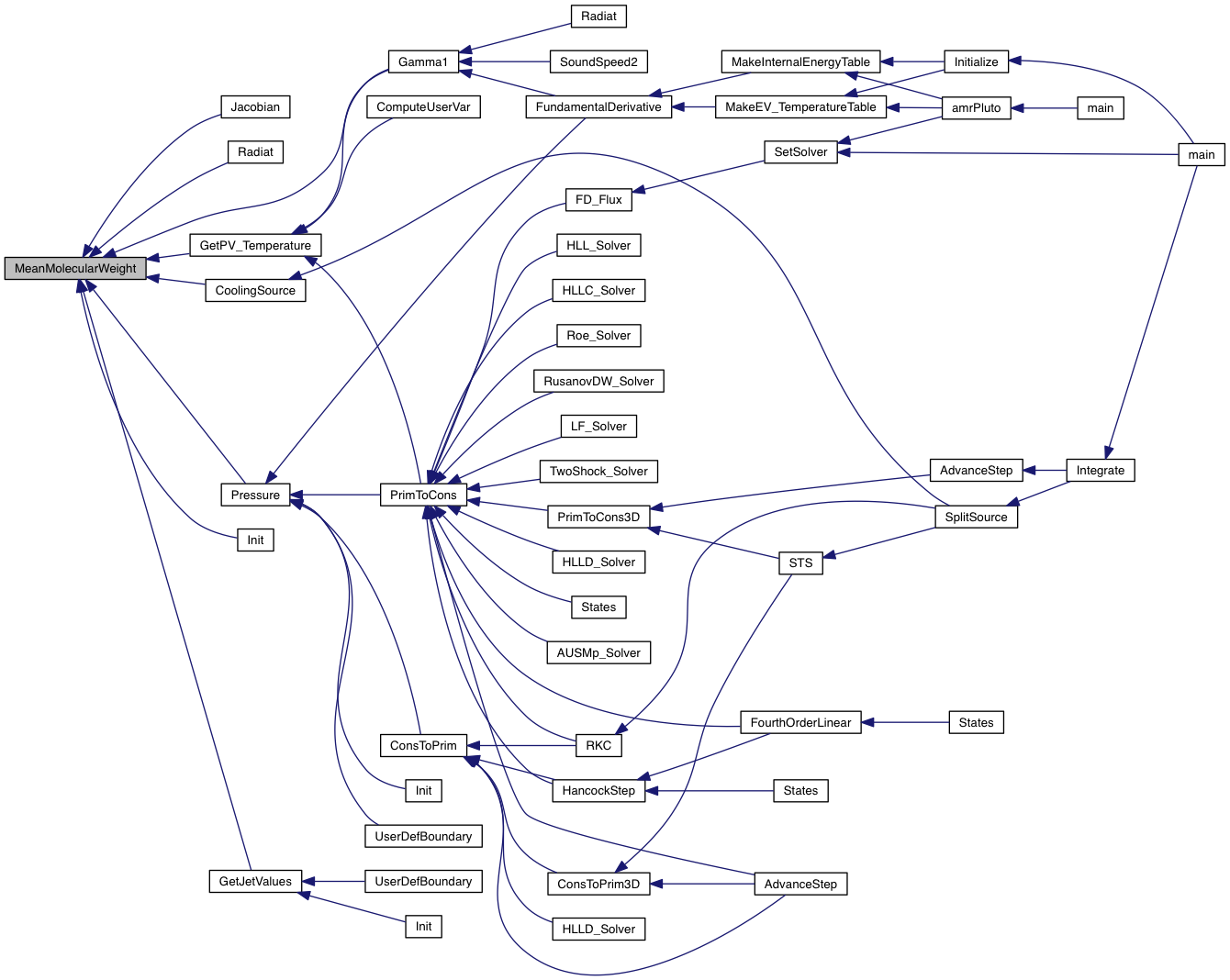
| double MeanMolecularWeight | ( | double * | v | ) |
Return the mean molecular weight.
| [in] | v | array of primitive variables (including ions) |
Definition at line 178 of file radiat.modified_by_Bhargav.c.

| double Median | ( | double | a, |
| double | b, | ||
| double | c | ||
| ) |
| FILE* OpenBinaryFile | ( | char * | filename, |
| int | sz, | ||
| char * | mode | ||
| ) |
Open a file for write/reading in binary mode.
| [in] | filename | a valid file name |
| [in] | sz | the distributed array descriptor. This parameter replaces dsize in parallel mode |
| [in] | mode | a string giving the opening mode (only "w" or "r" are allowed) |
Definition at line 31 of file bin_io.c.
| void OutflowBound | ( | double *** | q, |
| int | side, | ||
| int | vpos, | ||
| Grid * | grid | ||
| ) |
Impose zero-gradient boundary conditions on 'q' on the boundary side specified by box->side. The staggered component of magnetic field is assigned using the div.B = 0 condition in a different loop.
| [in,out] | q | a 3D array representing a flow variable |
| [in] | box | pointer to a RBox structure defining the extent of the boundary region and the variable position inside the cell |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 295 of file boundary.c.
| void ParabolicFlux | ( | Data_Arr | V, |
| Data_Arr | J, | ||
| double *** | T, | ||
| const State_1D * | state, | ||
| double ** | dcoeff, | ||
| int | beg, | ||
| int | end, | ||
| Grid * | grid | ||
| ) |
Add the diffusion fluxes to the upwind fluxes for explicit time integration.
| [in] | V | pointer to the 3D array of cell-centered primitive variables |
| [in,out] | state | pointer to a State_1D structure |
| [out] | dcoeff | the diffusion coefficients 1D array |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 34 of file parabolic_flux.c.
| [in] | V | 3D array containing primitive variables |
| [out] | dU | 3D array containing the conservative right hand sides |
| [in] | dt | the time step |
| [in] | grid | a pointer to an array of Grid structure |
Definition at line 50 of file parabolic_rhs.c.
| int ParamExist | ( | const char * | label | ) |
Check whether *label exists in any of the lines (**fline).
| [in] | label |
Definition at line 159 of file parse_file.c.
| char* ParamFileGet | ( | const char * | label, |
| int | pos | ||
| ) |
Search for *label in all the lines pointed to by **fline. If label exists, return a pointer to the string located pos words after label. Issue an error if this string cannot be located.
| [in] | label | the first word of a line to be searched |
| [in] | pos | an integer giving the position of the word |
Definition at line 86 of file parse_file.c.
| int ParamFileHasBoth | ( | const char * | label1, |
| const char * | label2 | ||
| ) |
Locate the line beginning with label1 and return 1 if label2 can be found on the same line. Return 0 otherwise.
| [in] | label1 | The first word of the line to be searched |
| [in] | label2 | A word containined in the same line beginning with label1 |
Definition at line 129 of file parse_file.c.

| int ParamFileRead | ( | char * | fname | ) |
Parse file *fname and store its content line by line in *fline. Blank lines are excluded.
| [in] | fname | the name of the file to be read |
Definition at line 53 of file parse_file.c.
| void ParseCmdLineArgs | ( | int | argc, |
| char * | argv[], | ||
| char * | ini_file, | ||
| Cmd_Line * | cmd | ||
| ) |
Parse command line options. Error messages will be output to stdout.
| [in] | argc | argument count |
| [in] | argv | argument vector |
| [in] | ini_file | a pointer to a string containing the name of the initialization file (default: "pluto.ini") |
| [out] | cmd | the command-line structure. |
Definition at line 18 of file cmd_line_opt.c.
| void PeriodicBound | ( | double *** | q, |
| int | side, | ||
| int | vpos | ||
| ) |
Implements periodic boundary conditions in serial mode.
Definition at line 568 of file boundary.c.
| void PlutoError | ( | int | condition, |
| char * | str | ||
| ) |
| void PrimToChar | ( | double ** | Lp, |
| double * | v, | ||
| double * | w | ||
| ) |
Compute the matrix-vector multiplcation between the the L matrix (containing primitive left eigenvectors) and the vector v. The result containing the characteristic variables is stored in the vector w = L.v
For efficiency purpose, multiplication is done explicitly, so that only nonzero entries of the left primitive eigenvectors are considered.
| [in] | L | Left eigenvectors |
| [in] | v | (difference of) primitive variables |
| [out] | w | (difference of) characteristic variables |
Compute the matrix-vector multiplcation between the the L matrix (containing primitive left eigenvectors) and the vector v. The result containing the characteristic variables is stored in the vector w = L.v
For efficiency purpose, multiplication is done explicitly, so that only nonzero entries of the left primitive eigenvectors are considered.
| [in] | Lp | Left eigenvectors |
| [in] | v | (difference of) primitive variables |
| [out] | w | (difference of) characteristic variables |
Definition at line 593 of file eigenv.c.
Convert a 3D array of primitive variables V to an array of conservative variables U. Note that [nv] is the fastest running index for U while it is the slowest running index for V.
| [in] | V | pointer to 3D array of primitive variables, with array indexing [nv][k][j][i] |
| [out] | U | pointer to 3D array of conserved variables, with array indexing [k][j][i][nv] |
| [in] | box | pointer to RBox structure containing the domain portion over which conversion must be performed. |
Definition at line 86 of file mappers3D.c.
| void print | ( | const char * | fmt, |
| ... | |||
| ) |
Define print function for the static grid version of PLUTO. The Chombo version is defined in Chombo/amrPLUTO.cpp
Definition at line 497 of file amrPluto.cpp.
| void print1 | ( | const char * | fmt, |
| ... | |||
| ) |
| void ReadBinaryArray | ( | void * | V, |
| size_t | dsize, | ||
| int | sz, | ||
| FILE * | fl, | ||
| int | istag, | ||
| int | swap_endian | ||
| ) |
Read a double-precision array from binary file.
| [in] | V | pointer to a 3D array, V[k][j][i] –> V[i + NX1*j + NX1*NX2*k]. Must start with index 0 |
| [in] | dsize | the size of the each buffer element (sizeof(double) or sizeof(float)). This parameter is used only in serial mode. |
| [in] | sz | the distributed array descriptor. This parameter replaces dsize in parallel mode |
| [in] | fl | a valid FILE pointer |
| [in] | istag | a flag to identify cell-centered (istag = -1) or staggered field data (istag = 0,1 or 2 for staggering in the x1, x2 or x3 directions) |
| [in] | swap_endian | a flag for swapping endianity |
Definition at line 143 of file bin_io.c.


Read data from disk using hdf5 format (double precision).
| [in] | output | the output structure associated with HDF5 format |
| [in] | grid | a pointer to an array of Grid structures |
Definition at line 582 of file hdf5_io.c.

| void ReflectiveBound | ( | double *** | q, |
| int | s, | ||
| int | side, | ||
| int | vpos | ||
| ) |
Make symmetric (s = 1) or anti-symmetric (s=-1) profiles with respect to the boundary plane specified by box->side. The sign is set by the FlipSign() function.
| [in,out] | q | a 3D flow quantity |
| [in] | box | pointer to box structure |
| [in] | s | an integer taking only the values +1 (symmetric profile) or -1 (antisymmetric profile) |
Definition at line 501 of file boundary.c.
Initialize some of the elements of the State_1D structure to zero in order to speed up computations. These includes:
| [in] | d | pointer to Data structure |
| [out] | state | pointer to a State_1D structure |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 163 of file set_indexes.c.
| void RestartDump | ( | Runtime * | ini | ) |
Write restart information needed for later restarts.
Definition at line 251 of file restart.c.

Read input binary / hdf5 data.
| [in] | ini | |
| [in] | nrestart | |
| [in] | type | specifies the output data type (type should be either DBL_OUTPUT or DBL_H5_OUTPUT). |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 17 of file restart.c.

| void RestartGet | ( | Runtime * | ini, |
| int | nrestart, | ||
| int | out_type, | ||
| int | swap_endian | ||
| ) |
Collect restart information needed for (potential) later restarts.
Definition at line 177 of file restart.c.

| void RightHandSide | ( | const State_1D * | state, |
| Time_Step * | Dts, | ||
| int | beg, | ||
| int | end, | ||
| double | dt, | ||
| Grid * | grid | ||
| ) |
| [in,out] | state | pointer to State_1D structure |
| [in] | Dts | pointer to time step structure |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
| [in] | dt | time increment |
| [in] | grid | pointer to Grid structure |
Definition at line 88 of file rhs.c.
| void RightHandSideSource | ( | const State_1D * | state, |
| Time_Step * | Dts, | ||
| int | beg, | ||
| int | end, | ||
| double | dt, | ||
| double * | phi_p, | ||
| Grid * | grid | ||
| ) |
| [in,out] | state | pointer to State_1D structure |
| [in] | Dts | pointer to time step structure |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
| [in] | dt | time increment |
| [in] | phi_p | force potential at interfaces |
| [in] | grid | pointer to Grid structure |
Definition at line 68 of file rhs_source.c.

Solve diffusion equation using Runge-Kutta-Chebyshev (RKC) method
| [in,out] | d | pointer to Data structure |
| [in,out] | Dts | pointer to Time_Step structure |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 58 of file rkc.c.

| Runtime* RuntimeGet | ( | void | ) |
| void RuntimeSet | ( | Runtime * | runtime | ) |
Store a static copy of the runtime structure for later access.
Definition at line 491 of file runtime_setup.c.
Open and parse the runtime initialization file. Assign values to the runtime structure.
| [out] | runtime | pointer to a Runtime structure |
| [in] | cmd_line | pointer to a Cmd_Line structure (useful, e.g., to resize the domain using the -xres option) |
| [in] | ini_file | the name of the initialization file (default is "pluto.ini") specified with the -i option. |
Definition at line 22 of file runtime_setup.c.
| void SetColorMap | ( | unsigned char * | , |
| unsigned char * | , | ||
| unsigned char * | , | ||
| char * | |||
| ) |
Definition at line 11 of file colortable.c.

| void SetDefaultVarNames | ( | Output * | ) |
Definition at line 4 of file var_names.c.

| int SetDumpVar | ( | char * | var_name, |
| int | out_type, | ||
| int | flag | ||
| ) |
Include ('flag == YES') or exclude ('flag == NO') the variable associated to 'var_name' in or from the output type 'out_type'. If 'out_type' corresponds to an image (ppm or png), create a correspdonding Image structure.
| [in] | var_name | the name of the variable (e.g. "rho", "vx1",...) |
| [in] | out_type | select the output type (e.g., DBL_OUTPUT, VTK_OUTPUT, and so forth) |
| [in] | flag | an integer values (YES/NO). |
Definition at line 211 of file set_output.c.

| [in] | rtime | pointer to a Runtime structure |
| [out] | GXYZ | pointer to array of Grid structures |
Definition at line 21 of file set_grid.c.
Set vector indices and integration index range.
| [out] | indx | pointer to an Index structure |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 49 of file set_indexes.c.

Adjust the size of computational domain by reducing the final computationa index in the direction of propagation.
| [in] | d | pointer to Data structure |
| [in] | dir | the direction of propagation |
| [in] | log_freq | the output log frequency |
| [in,out] | grid | pointer to array of Grid structures |
Definition at line 29 of file jet_domain.c.

| int SetLogFile | ( | char * | output_dir, |
| Cmd_Line * | cmd | ||
| ) |
Set default attributes (variable names, pointers to data structures, filename extensions, etc...) of the output structures.
| [in] | d | pointer to Data structure |
| [in] | runtime | pointer to Runtime structure |
Definition at line 37 of file set_output.c.

| void SetRBox | ( | void | ) |
Definition at line 33 of file rbox.c.
| Riemann_Solver* SetSolver | ( | const char * | solver | ) |
PURPOSE
return a pointer to a riemann solver function
Depending on the choice of the Riemann solver specified in pluto.ini, return a pointer to the corresponding Riemann solver function
Definition at line 4 of file set_solver.c.
| void Show | ( | double ** | a, |
| int | ip | ||
| ) |
| void ShowConfig | ( | int | argc, |
| char * | argv[], | ||
| char * | ini_file | ||
| ) |
Write a summary of the selected options
/ _ \/ / / / / /_ __/ __ \ / ___/ /__/ /_/ / / / / /_/ /
Definition at line 16 of file show_config.c.
| void ShowDomainDecomposition | ( | int | nprocs, |
| Grid * | GXYZ | ||
| ) |
Show the parallel domain decomposition by having each processor print its own computational domain. This is activated with the -show-dec command line argument. It may be long for several thousand processors.
| [in] | nprocs | the total number of processors |
| [in] | GXYZ | a pointer to an array of grid structures |
Definition at line 295 of file show_config.c.

| void ShowMatrix | ( | double ** | A, |
| int | n, | ||
| double | eps | ||
| ) |
| void ShowUnits | ( | ) |
Show units when cooling is enabled.
Definition at line 260 of file show_config.c.
| void ShowVector | ( | double * | v, |
| int | n | ||
| ) |
| void SoundSpeed2 | ( | double ** | v, |
| double * | cs2, | ||
| double * | h, | ||
| int | beg, | ||
| int | end, | ||
| int | pos, | ||
| Grid * | grid | ||
| ) |
Define the square of the sound speed.
| [in] | v | 1D array of primitive quantities |
| [out] | cs2 | 1D array containing the square of the sound speed |
| [in] | h | 1D array of enthalpy values |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
| [in] | pos | an integer specifying the spatial position inside the cell (only for spatially-dependent EOS) |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 16 of file eos.c.
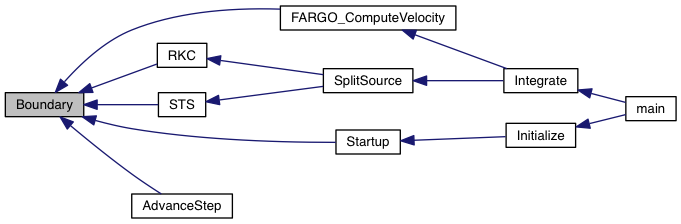
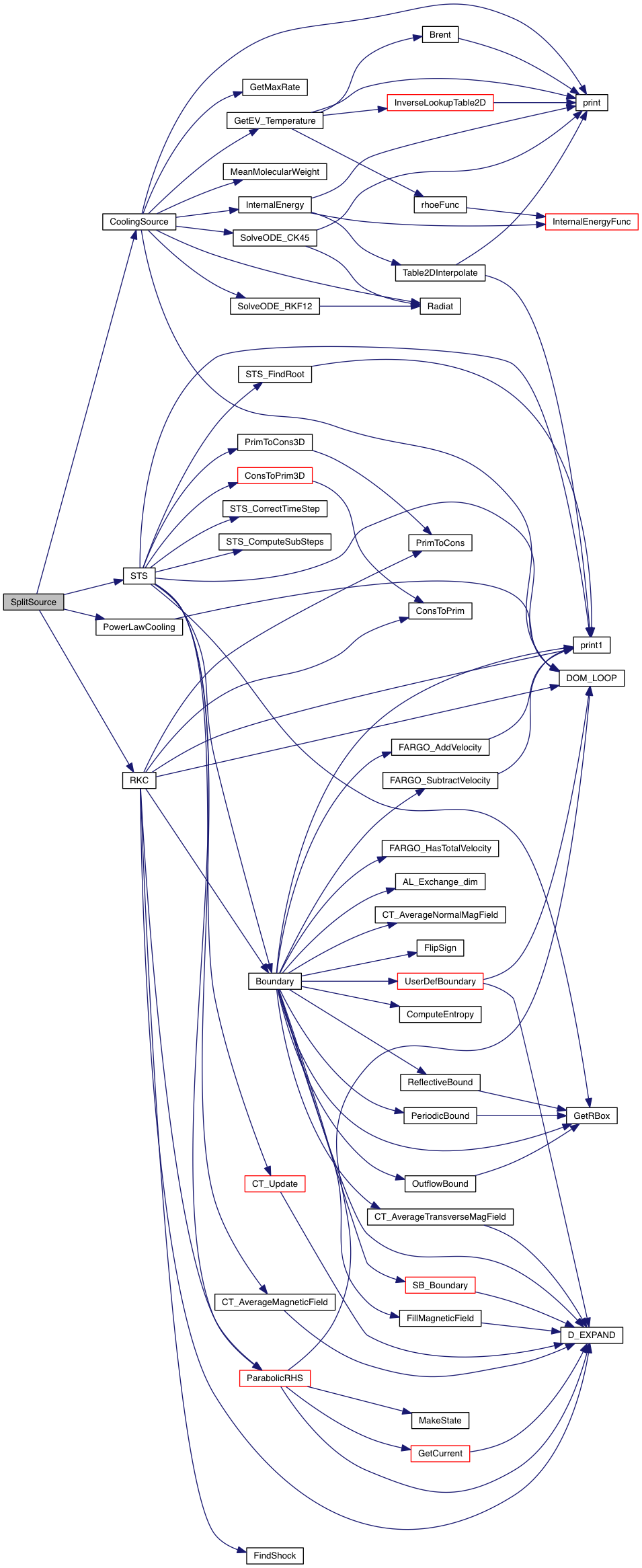
Take one step on operator-split source terms.
| [in,out] | d | pointer to PLUTO Data structure containing the solution array updated from the most recent call |
| [in] | dt | the time step used to integrate the source terms |
| [in] | Dts | pointer to the time step structure |
| [in] | grid | pointer to an array of grid structures |
Definition at line 25 of file split_source.c.

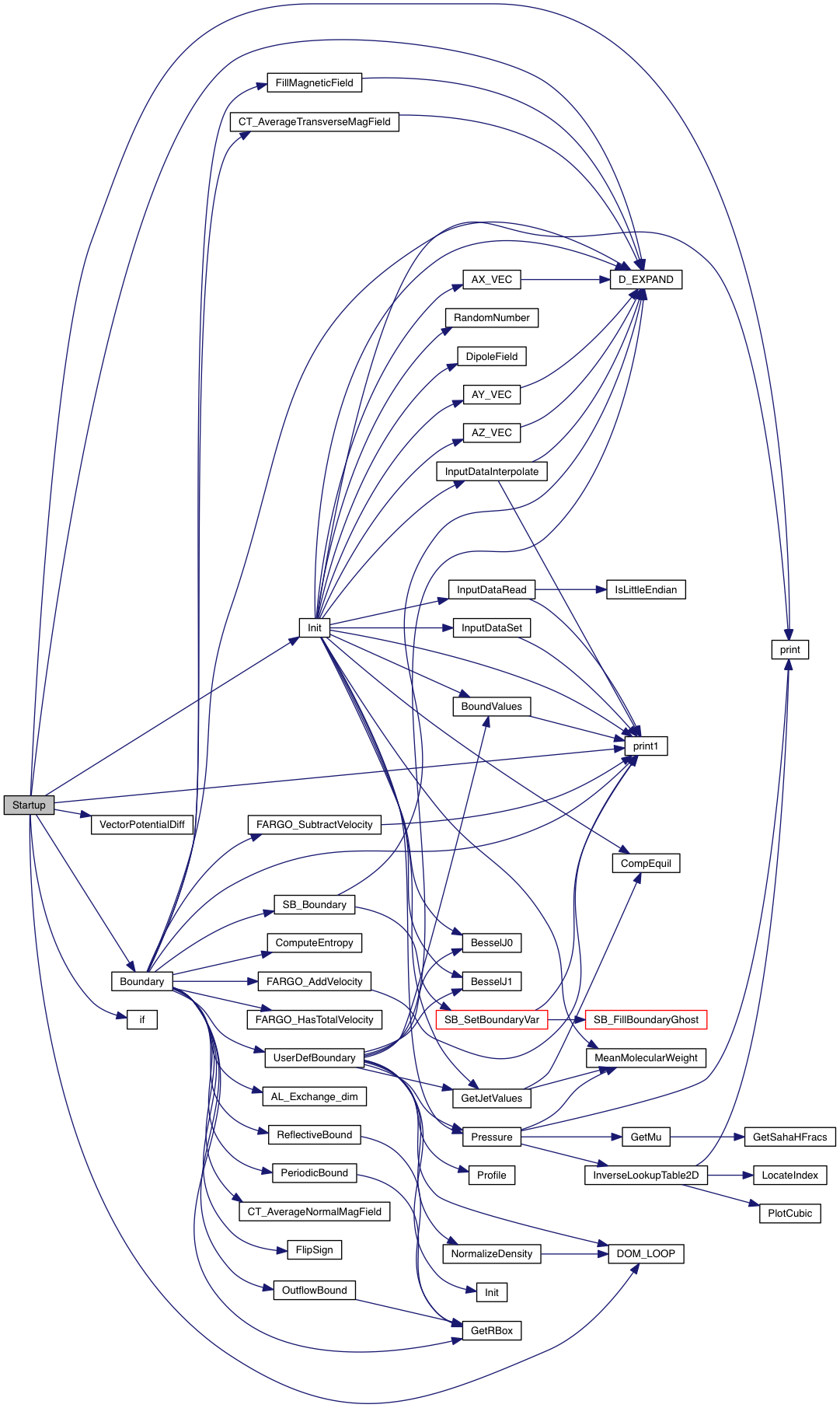
Definition at line 17 of file startup.c.

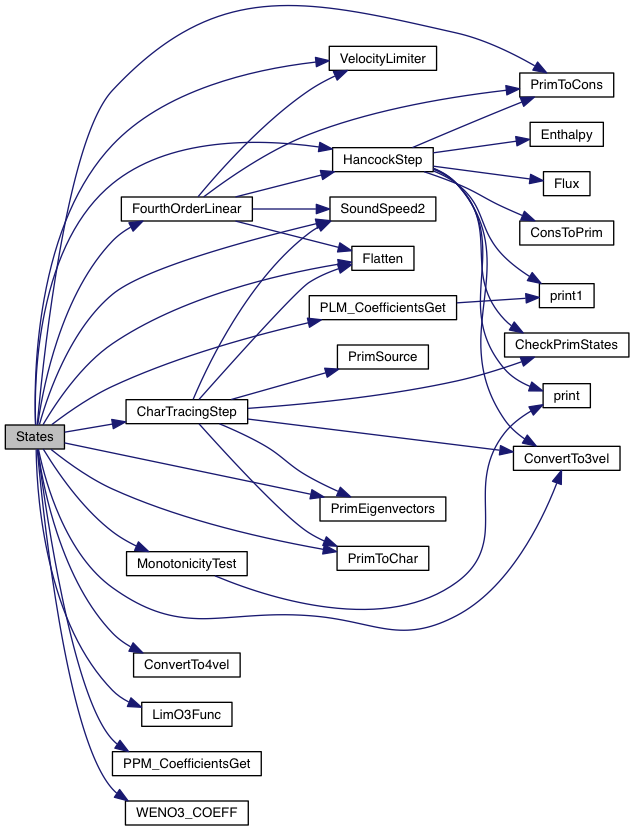
Compute 1D left and right interface states using piecewise linear reconstruction and the characteristic decomposition of the quasi-linear form of the equations.
This is done by first extrapolating the cell center value to the interface using piecewise limited linear reconstruction on the characteristic variables.
Left and right states are then evolved for the half time step using characteristic tracing if necessary.
Compute states using piecewise linear interpolation.
| [in] | state | pointer to a State_1D structure |
| [in] | beg | starting point where vp and vm must be computed |
| [in] | end | final point where vp and vm must be computed |
| [in] | grid | pointer to array of Grid structures |
Compute 1D left and right interface states using piecewise linear reconstruction and the characteristic decomposition of the quasi-linear form of the equations.
This is done by first extrapolating the cell center value to the interface using piecewise limited linear reconstruction on the characteristic variables.
Left and right states are then evolved for the half time step using characteristic tracing if necessary.
| [in] | state | pointer to State_1D structure |
| [in] | beg | initial index of computation |
| [in] | end | final index of computation |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 430 of file plm_states.c.
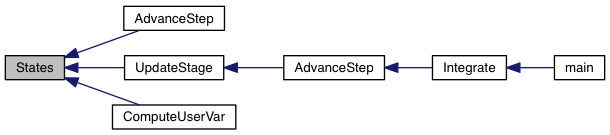
Solve diffusion equation(s) using Super-Time-Stepping.
| [in,out] | d | pointer to Data structure |
| [in,out] | Dts | pointer to Time_Step structure |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 55 of file sts.c.

| void SwapEndian | ( | void * | x, |
| const int | nbytes | ||
| ) |
| void Trace | ( | double | xx | ) |
Restore original (full domain) indexes.
Definition at line 154 of file jet_domain.c.

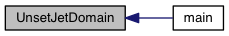
| void UpdateStage | ( | const Data * | d, |
| Data_Arr | UU, | ||
| double ** | aflux, | ||
| Riemann_Solver * | Riemann, | ||
| double | dt, | ||
| Time_Step * | Dts, | ||
| Grid * | grid | ||
| ) |
| [in,out] | d | pointer to PLUTO Data structure |
| [in,out] | UU | data array containing conservative variables at the previous time step to be updated |
| [out] | aflux | interface fluxes needed for refluxing operations (only with AMR) |
| [in] | Riemann | pointer to a Riemann solver function |
| [in] | dt | the time step for the current update step |
| [in,out] | Dts | pointer to time step structure |
| [in] | grid | pointer to array of Grid structures |
Definition at line 38 of file update_stage.c.

Assign user-defined boundary conditions.
| [in,out] | d | pointer to the PLUTO data structure containing cell-centered primitive quantities (d->Vc) and staggered magnetic fields (d->Vs, when used) to be filled. |
| [in] | box | pointer to a RBox structure containing the lower and upper indices of the ghost zone-centers/nodes or edges at which data values should be assigned. |
| [in] | side | specifies the boundary side where ghost zones need to be filled. It can assume the following pre-definite values: X1_BEG, X1_END, X2_BEG, X2_END, X3_BEG, X3_END. The special value side == 0 is used to control a region inside the computational domain. |
| [in] | grid | pointer to an array of Grid structures. |
Assign user-defined boundary conditions in the lower boundary ghost zones. The profile is top-hat:
![\[ V_{ij} = \left\{\begin{array}{ll} V_{\rm jet} & \quad\mathrm{for}\quad r_i < 1 \\ \noalign{\medskip} \mathrm{Reflect}(V) & \quad\mathrm{otherwise} \end{array}\right. \]](form_235.png)
where 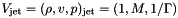 and
and M is the flow Mach number (the unit velocity is the jet sound speed, so 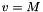 ).
).
Assign user-defined boundary conditions:
x < 1/6 and reflective boundary otherwise.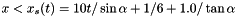 we use fixed (post-shock) values. Unperturbed values otherwise.
we use fixed (post-shock) values. Unperturbed values otherwise.Assign user-defined boundary conditions at inner and outer radial boundaries. Reflective conditions are applied except for the azimuthal velocity which is fixed.
Assign user-defined boundary conditions.
| [in/out] | d pointer to the PLUTO data structure containing cell-centered primitive quantities (d->Vc) and staggered magnetic fields (d->Vs, when used) to be filled. | |
| [in] | box | pointer to a RBox structure containing the lower and upper indices of the ghost zone-centers/nodes or edges at which data values should be assigned. |
| [in] | side | specifies on which side boundary conditions need to be assigned. side can assume the following pre-definite values: X1_BEG, X1_END, X2_BEG, X2_END, X3_BEG, X3_END. The special value side == 0 is used to control a region inside the computational domain. |
| [in] | grid | pointer to an array of Grid structures. |
Set the injection boundary condition at the lower z-boundary (X2-beg must be set to userdef in pluto.ini). For 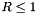 we set constant input values (given by the GetJetValues() function while for $ R > 1 $ the solution has equatorial symmetry with respect to the
we set constant input values (given by the GetJetValues() function while for $ R > 1 $ the solution has equatorial symmetry with respect to the z=0 plane. To avoid numerical problems with a "top-hat" discontinuous jump, we smoothly merge the inlet and reflected value using a profile function Profile().
Provide inner radial boundary condition in polar geometry. Zero gradient is prescribed on density, pressure and magnetic field. For the velocity, zero gradient is imposed on v/r (v = vr, vphi).
The user-defined boundary is used to impose stress-free boundary and purely vertical magnetic field at the top and bottom boundaries, as done in Bodo et al. (2012). In addition, constant temperature and hydrostatic balance are imposed. For instance, at the bottom boundary, one has:
![\[ \left\{\begin{array}{lcl} \DS \frac{dp}{dz} &=& \rho g_z \\ \noalign{\medskip} p &=& \rho c_s^2 \end{array}\right. \qquad\Longrightarrow\qquad \frac{p_{k+1}-p_k}{\Delta z} = \frac{p_{k} + p_{k+1}}{2c_s^2}g_z \]](form_395.png)
where 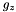 is the value of gravity at the lower boundary. Solving for
is the value of gravity at the lower boundary. Solving for 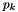 at the bottom boundary where
at the bottom boundary where 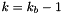 gives:
gives:
![\[ \left\{\begin{array}{lcl} p_k &=& \DS p_{k+1} \frac{1-a}{1+a} \\ \noalign{\medskip} \rho_k &=& \DS \frac{p_k}{c_s^2} \end{array}\right. \qquad\mathrm{where}\qquad a = \frac{\Delta z g_z}{2c_s^2} > 0 \]](form_399.png)
where, for simplicity, we keep constant temperature in the ghost zones rather than at the boundary interface (this seems to give a more stable behavior and avoids negative densities). A similar treatment holds at the top boundary.
Assign user-defined boundary conditions. At the inner boundary we use outflow conditions, except for velocity which we reset to zero when there's an inflow
Definition at line 98 of file init.c.
| void VectorPotentialDiff | ( | double * | , |
| int | , | ||
| int | , | ||
| int | , | ||
| Grid * | |||
| ) |

| void VectorPotentialUpdate | ( | const Data * | d, |
| const void * | vp, | ||
| const State_1D * | state, | ||
| const Grid * | grid | ||
| ) |
| void Where | ( | int | i, |
| Grid * | grid | ||
| ) |
Print the location of a particular zone (i,j,k) in the computational domain.
Definition at line 235 of file tools.c.
| void WriteAsciiFile | ( | char * | fname, |
| double * | q, | ||
| int | nvar | ||
| ) |
| void WriteBinaryArray | ( | void * | V, |
| size_t | dsize, | ||
| int | sz, | ||
| FILE * | fl, | ||
| int | istag | ||
| ) |
Write an array to disk in binary format.
| [in] | V | single pointer to a 3D array, V[k][j][i] –> V[i + NX1*j + NX1*NX2*k]. Must start with index 0 |
| [in] | dsize | the size of the each buffer element (sizeof(double) or sizeof(float)). This parameter is used only in serial mode. |
| [in] | sz | the distributed array descriptor. This parameter replaces dsize in parallel mode |
| [in] | fl | a valid FILE pointer |
| [in] | istag | a flag to identify cell-centered (istag = -1) or staggered field data (istag = 0,1 or 2 for staggering in the x1, x2 or x3 directions) |
Definition at line 98 of file bin_io.c.

Write data to disk using any of the available formats.
| [in] | d | pointer to PLUTO Data structre |
| [in] | output | the output structure corresponding to a given format |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 29 of file write_data.c.

Write data to disk using hdf5 format in single or double precision.
| [in] | output | the output structure associated with HDF5 format |
| [in] | grid | a pointer to an array of Grid structures |
Definition at line 45 of file hdf5_io.c.

| void WritePNG | ( | double *** | , |
| char * | , | ||
| char * | , | ||
| Grid * | |||
| ) |

| void WritePPM | ( | double *** | , |
| char * | , | ||
| char * | , | ||
| Grid * | |||
| ) |
Definition at line 10 of file write_img.c.

Write tabulated array.
| [in] | output | a pointer to the output structure corresponding to the tab format |
| [in] | filename | the output file name |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 31 of file write_tab.c.

| void WriteVTK_Header | ( | FILE * | fvtk, |
| Grid * | grid | ||
| ) |
Write VTK header in parallel or serial mode. In parallel mode only processor 0 does the actual writing (see al_io.c/AL_Write_header).
| [in] | fvtk | pointer to file |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 91 of file write_vtk.c.

| void WriteVTK_Scalar | ( | FILE * | fvtk, |
| double *** | V, | ||
| double | unit, | ||
| char * | var_name, | ||
| Grid * | grid | ||
| ) |
Write VTK scalar field.
| [in] | fvtk | pointer to file (handle) |
| [in] | V | pointer to 3D data array |
| [in] | unit | the corresponding cgs unit (if specified, 1 otherwise) |
| [in] | var_name | the variable name appearing in the VTK file |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 341 of file write_vtk.c.

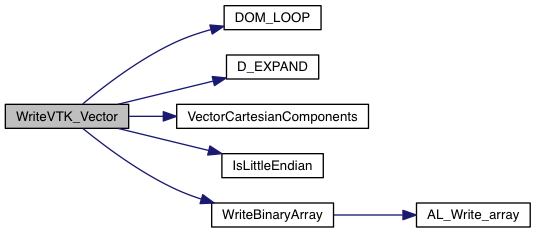
Write VTK vector field data. This is enabled only when VTK_VECTOR_DUMP is set to YES. For generality purposes, vectors are written always with 3 components, even when there're only 2 being used.
The following Maple script has been used to find vector components from cyl/sph to cartesian:
| [in] | fvtk | pointer to file |
| [in] | V | a 4D array [nv][k][j][i] containing the vector components (nv) defined at cell centers (k,j,i). The index nv = 0 marks the vector first component. |
| [in] | unit | the corresponding cgs unit (if specified, 1 otherwise) |
| [in] | var_name | the variable name appearing in the VTK file |
| [in] | grid | pointer to an array of Grid structures |
Definition at line 259 of file write_vtk.c.