|
PLUTO
|
|
PLUTO
|
Disk-Planet interaction problem. More...
#include "pluto.h"
Go to the source code of this file.
Macros | |
| #define | MIN_DENSITY 1e-8 |
| #define | g_OmegaZ 0.0 |
Functions | |
| static void | NormalizeDensity (const Data *d, Grid *g) |
| void | Init (double *us, double x1, double x2, double x3) |
| void | Analysis (const Data *d, Grid *grid) |
| void | UserDefBoundary (const Data *d, RBox *box, int side, Grid *grid) |
Disk-Planet interaction problem.
Simulate the interaction of a planet embedded in a disk as described in section 3.4 of Mignone et al., A&A (2012) 545, A152. This test is a nice benchmark for the FARGO module and the ROTATING_FRAME switch. For testing-purposes no viscosity is used here. The initial condition consists of a locally isothermal configuration with temperature profile 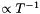 yielding a disk vertical height to radius of
yielding a disk vertical height to radius of 0.05. The gravitational potential due to the presence of the star and the planet is defined in BodyForcePotential() function.
The conventions used throught the implementation are the following:
r = spherical radiusR = cylindrical radiusz = cylindrical heightth = meridional angleThe test can be carried out in polar (2D or 3D) or spherical (3D) coordinates and the following parameters determine the initial configuration:
g_inputParam[Mstar]: controls the star mass (in solar masses)g_inputParam[Mdisk]: controls the disk mass (in solar masses)g_inputParam[Mplanet]: sets the planet mass (in earth masses)g_inputParam[Viscosity]: sets the amount of viscosityComputation can be carried in the rotating or in the observer's frame of reference (ROTATING_FRAME to YES or NO, respectively). In particular:

References:
Definition in file init.c.
| void Init | ( | double * | us, |
| double | x1, | ||
| double | x2, | ||
| double | x3 | ||
| ) |
The Init() function can be used to assign initial conditions as as a function of spatial position.
| [out] | v | a pointer to a vector of primitive variables |
| [in] | x1 | coordinate point in the 1st dimension |
| [in] | x2 | coordinate point in the 2nd dimension |
| [in] | x3 | coordinate point in the 3rdt dimension |
The meaning of x1, x2 and x3 depends on the geometry:
![\[ \begin{array}{cccl} x_1 & x_2 & x_3 & \mathrm{Geometry} \\ \noalign{\medskip} \hline x & y & z & \mathrm{Cartesian} \\ \noalign{\medskip} R & z & - & \mathrm{cylindrical} \\ \noalign{\medskip} R & \phi & z & \mathrm{polar} \\ \noalign{\medskip} r & \theta & \phi & \mathrm{spherical} \end{array} \]](form_173.png)
Variable names are accessed by means of an index v[nv], where nv = RHO is density, nv = PRS is pressure, nv = (VX1, VX2, VX3) are the three components of velocity, and so forth.
Definition at line 68 of file init.c.
Definition at line 199 of file init.c.

Assign user-defined boundary conditions.
| [in,out] | d | pointer to the PLUTO data structure containing cell-centered primitive quantities (d->Vc) and staggered magnetic fields (d->Vs, when used) to be filled. |
| [in] | box | pointer to a RBox structure containing the lower and upper indices of the ghost zone-centers/nodes or edges at which data values should be assigned. |
| [in] | side | specifies the boundary side where ghost zones need to be filled. It can assume the following pre-definite values: X1_BEG, X1_END, X2_BEG, X2_END, X3_BEG, X3_END. The special value side == 0 is used to control a region inside the computational domain. |
| [in] | grid | pointer to an array of Grid structures. |
Definition at line 137 of file init.c.
