|
PLUTO
|
|
PLUTO
|
Integrate cooling and reaction source terms. More...
#include "pluto.h"
Go to the source code of this file.
Functions | |
| void | CoolingSource (const Data *d, double dt, Time_Step *Dts, Grid *GXYZ) |
| void | Numerical_Jacobian (double *v, double **J) |
Integrate cooling and reaction source terms.
Solve the system of ordinary differential equations describing optically thin cooling and ionization network (if any). We use an adaptive step size integration method and follow the strategy outlined in section 5.1 of Tesileanu et al. (2008) for handling stiff cells.
On output, a time-step estimate for the next time level is computed using the relative or absolute variation obtained during the integration of the ODE (from t(n) –> t(n+1))
![\[ \Delta t_c = \min_{ijk}\left[\frac{\Delta t^n M_R}{\epsilon}\right] \,\quad\rm{where}\quad \epsilon = \max\left(\left|\frac{p^{n+1}}{p^n} - 1\right|,\, |X^{n+1}-X^n|\right) \]](form_5.png)
where 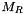 is the maximum cooling rate (defined by the global variable g_maxCoolingRate) and X are the chemical species.
is the maximum cooling rate (defined by the global variable g_maxCoolingRate) and X are the chemical species.
References
Definition in file cooling_source.c.
Integrate cooling and reaction source terms.
| [in,out] | d | pointer to Data structure |
| [in] | dt | the time step to be taken |
| [out] | Dts | pointer to the Time_Step structure |
| [in] | GXYZ | pointer to an array of Grid structures |
Definition at line 38 of file cooling_source.c.

| void Numerical_Jacobian | ( | double * | v, |
| double ** | J | ||
| ) |
Compute Jacobian matrix numerically and get eigenvector and eigenvalue decomposition
The purpose of this function is to detect stiffness.
Note: This function is EXTREMELY time consuming.
Definition at line 227 of file cooling_source.c.
