143 double dBx, dpsi, dtdx, dt, dtdV;
144 double *
A, *
dx, *
dV, r;
145 double *vp, *vm, *vc, dvp[
NVAR], dvm[
NVAR];
148 static double **
fp, **fm, **uh, **u;
149 static double *pp, *pm, *hp, *hm;
150 static double *lambda_max, *lambda_min;
152 #if GEOMETRY != CARTESIAN && GEOMETRY != CYLINDRICAL
153 print1 (
"! Hancock does not work in this geometry \n");
157 print (
"! Conservative Hancock scheme does not support gravity\n");
160 #if RECONSTRUCTION != LINEAR
161 print1 (
"! The MUSCL-Hancock scheme works with Linear reconstruction only\n");
164 #if (PARABOLIC_FLUX & EXPLICIT)
165 print1 (
"! Conservative MUSCL-Hancock incompatible with Explicit Diffusion\n");
211 for (i =
beg; i <=
end; i++){
212 vp = state->
vp[
i]; vm = state->
vm[
i];
215 fp[
i][nv] = fp[
i][
RHO]*vp[nv];
216 fm[
i][nv] = fm[
i][
RHO]*vm[nv];
222 #if CHAR_UPWIND == YES
223 MAX_CH_SPEED (state->
v, lambda_min, lambda_max,
beg,
end);
231 for (i =
beg; i <=
end; i++) {
236 #if GEOMETRY == CARTESIAN
237 for (nv =
NVAR; nv--; ) rhs[i][nv] = -dtdx*(fp[i][nv] - fm[i][nv]);
238 #elif GEOMETRY == CYLINDRICAL
240 for (nv =
NVAR; nv--; ) {
241 rhs[
i][nv] = -dtdV*(A[
i]*fp[
i][nv] - A[i-1]*fm[
i][nv]);
247 rhs[
i][
MXn] -= dtdx*(pp[
i] - pm[
i]);
251 #if (PHYSICS == MHD || PHYSICS == RMHD) && (DIVB_CONTROL == EIGHT_WAVES)
252 dBx = (vp[
BXn]*A[
i] - vm[
BXn]*A[i-1])/dV[i];
253 EXPAND(rhs[i][
BX1] -= dt*state->
v[i][
VX1]*dBx; ,
254 rhs[i][
BX2] -= dt*state->
v[i][
VX2]*dBx; ,
255 rhs[i][
BX3] -= dt*state->
v[i][
VX3]*dBx;)
261 #if GLM_EXTENDED == YES
262 dBx = (vp[
BXn]*A[
i] - vm[
BXn]*A[i-1])/dV[i];
263 EXPAND(rhs[i][
MX1] -= dt*state->
v[i][
BX1]*dBx; ,
264 rhs[i][
MX2] -= dt*state->
v[i][
BX2]*dBx; ,
265 rhs[i][
MX3] -= dt*state->
v[i][
BX3]*dBx;)
268 rhs[
i][ENG] -= dt*state->
v[
i][
BXn]*dpsi;
278 #if GEOMETRY == CYLINDRICAL && COMPONENTS == 3
280 double vB,
vel2, g_2,
r_1, *uc;
282 for (i =
beg; i <=
end; i++){
305 for (i =
beg; i <=
end; i++) {
306 for (nv =
NVAR; nv--; ) uh[i][nv] = u[i][nv] + rhs[i][nv];
315 for (i =
beg; i <=
end; i++){
316 if (state->
flag[i] != 0){
317 for (nv =
NVAR; nv--; ){
318 uh[
i][nv] = u[
i][nv];
319 state->
vh[
i][nv] = state->
v[
i][nv];
320 state->
vp[
i][nv] = state->
vm[
i][nv] = state->
v[
i][nv];
334 for (i =
beg; i <=
end; i++) {
339 for (nv =
NVAR; nv--; ){
350 P1 = vp[nv] - vm[nv];
351 dvp[nv] = dvm[nv] = state->
vh[
i][nv] - state->
v[
i][nv];
356 #if CHAR_UPWIND == YES
358 if (lambda_min[i] > 0.0)
359 for (nv =
NVAR; nv--; ) dvm[nv] = 0.0;
360 else if (lambda_max[i] < 0.0)
361 for (nv =
NVAR; nv--; ) dvp[nv] = 0.0;
365 for (nv =
NVAR; nv--; ){
366 vp[nv] = vc[nv] + dvp[nv];
367 vm[nv] = vc[nv] + dvm[nv];
371 for (i =
beg-1; i <=
end; i++){
double ** v
Cell-centered primitive varables at the base time level, v[i] = .
int end
Global end index for the local array.
double ** vh
Primitive state at n+1/2 (only for one step method)
void print1(const char *fmt,...)
void Flux(double **u, double **w, double *a2, double **fx, double *p, int beg, int end)
double ** rhs
Conservative right hand side.
void CheckPrimStates(double **vM, double **vP, double **v0, int beg, int end)
void Enthalpy(double **v, real *h, int beg, int end)
double g_dt
The current integration time step.
double ** vp
prim vars at i+1/2 edge, vp[i] = vL(i+1/2)
int g_dir
Specifies the current sweep or direction of integration.
int beg
Global start index for the local array.
int ConsToPrim(double **ucons, double **uprim, int ibeg, int iend, unsigned char *flag)
void ConvertTo3vel(double **, int, int)
void print(const char *fmt,...)
double * r_1
Geometrical factor 1/r.
double ** vm
prim vars at i-1/2 edge, vm[i] = vR(i-1/2)
#define ARRAY_1D(nx, type)
long int NMAX_POINT
Maximum number of points among the three directions, boundaries excluded.
void PrimToCons(double **uprim, double **ucons, int ibeg, int iend)
double * bn
Face magentic field, bn = bx(i+1/2)
double ** um
same as vm, in conservative vars
double ** up
same as vp, in conservative vars
#define ARRAY_2D(nx, ny, type)
#define QUIT_PLUTO(e_code)
double * A
Right interface area, A[i] = .
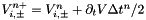
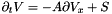 .
.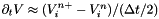 . This requires taking the following steps:
. This requires taking the following steps: